ExcelRT™ Reverse Transcription Kit II, 100 rx
ExcelRT™ Reverse Transcription Kit II adalah kit yang lengkap, efisien dan nyaman untuk mensintesis cDNA berkualitas tinggi. Kit ini memiliki ExcelRT™ Reverse Transcriptase, yang mampu mensintesis cDNA pada suhu 37~50°C. ExcelRT™ Reverse Transcriptase merupakan rekombinan Moloney Murine Leukemia Virus (M-MLV), yang dirancang untuk mengurangi aktivitas RNase H dan menciptakan stabilitas termal yang lebih baik. Kit ini juga mengandung RNAok™ RNase Inhibitor, yang aktif melawan RNase A, RNase B, dan RNase C. Produk ini dilengkapi dengan RT Buffer dan Oligo (dT)/Random Primer Mix yang dioptimalkan untuk sintesis cDNA yang sangat efisien dan cocok untuk penelitian PCR dan realtime PCR.
Spesifikasi Teknis
Setiap kit terdiri dari:
RTase/RI Enzyme Mix 100 μl
5X RT Buffer (DTT/dNTPs) 500 μl
Oligo (dT)/Random Primer Mix 100 μl
DEPC-Treated H2O, 1 ml x 2
Kapasitas penggunaan: 100 reaksi (@20ul/reaksi)
Penyimpanan
-20°C selama 24 bulan
Riwayat Publikasi:
Identification of co-regulated genes associated with doxorubicin resistance in the MCF-7/ADR cancer cell line
Ali Miri, Javad Gharechahi, Iman Samiei Mosleh, Kazem Sharifi, Vahid Jajarmi. Front Oncol. 2023; 13: 1135836. Published online 2023 Jun 15. doi: 10.3389/fonc.2023.1135836
PMCID: PMC10311417
Seyyed Morteza Hoseini, Morteza Yousefi, Alireza Afzali-Kordmahalleh, Esmaeil Pagheh, Ali Taheri Mirghaed
Animals (Basel) 2023 Jun; 13(12): 1934. Published online 2023 Jun 9. doi: 10.3390/ani13121934
PMCID: PMC10295004
Proline-related gene expression contribute to physiological changes of East Nusa Tenggara (Indonesia) local rice cultivars during drought stress
YUSTINA CAROLINA FEBRIANTI SALSINHA, SITI NURBAITI, ALFINO SEBASTIAN,DIDIK INDRADEWA, YEKTI ASIH PURWESTRI, DIAH RACHMAWATI, BIODIVERSITAS 2022 July Volume 23, Number 7, Pages: 3573-3583
Evolutionary dynamics of SARS-CoV-2 circulating in Yogyakarta and Central Java, Indonesia: sequence analysis covering furin cleavage site (FCS) region of the spike protein
Nastiti Wijayanti, Faris Muhammad Gazali, Endah Supriyati, Mohamad Saifudin Hakim, Eggi Arguni, Marselinus Edwin Widyanto Daniwijaya, Titik Nuryastuti, Matin Nuhamunada, Rahma Nabilla, Sofia Mubarika Haryana, Tri Wibawa, Int Microbiol. 2022 Feb 14;1-10. doi: 10.1007/s10123-022-00239-8.
PMCID: PMC8853438
Detection of SARS-CoV-2 spike protein D614G mutation by qPCR-HRM analysis
Faris Muhammad Gazali, Matin Nuhamunada, Rahma Nabilla, Endah Supriyati, Mohamad Saifudin Hakim, Eggi Arguni, Edwin Widyanto Daniwijaya, Titik Nuryastuti, Sofia Mubarika Haryana, Tri Wibawa, Nastiti Wijayanti, Heliyon. 2021 Sep;7(9):e07936. doi: 10.1016/j.heliyon.2021.e07936. Epub 2021 Sep 6.
PMCID: PMC8420086
BMPR2 Editing in Fibroblast NIH3T3 using CRISPR/Cas9 Affecting BMPR2 mRNA Expression and Proliferation
Dwi Aris Agung Nugrahaningsih, Eko Purnomo, Widya Wasityastuti, Ronny Martien, Nur Arfian, Tety Hartatik, The Indonesian Biomedical Journal, 2022, Vol 14, No 1. DOI: https://doi.org/10.18585/inabj.v14i1.1724
Spesifikasi Teknis
Setiap kit terdiri dari:
RTase/RI Enzyme Mix 100 μl
5X RT Buffer (DTT/dNTPs) 500 μl
Oligo (dT)/Random Primer Mix 100 μl
DEPC-Treated H2O, 1 ml x 2
Kapasitas penggunaan: 100 reaksi (@20ul/reaksi)
Penyimpanan
-20°C selama 24 bulan
Riwayat Publikasi:
Identification of co-regulated genes associated with doxorubicin resistance in the MCF-7/ADR cancer cell line
Ali Miri, Javad Gharechahi, Iman Samiei Mosleh, Kazem Sharifi, Vahid Jajarmi. Front Oncol. 2023; 13: 1135836. Published online 2023 Jun 15. doi: 10.3389/fonc.2023.1135836
PMCID: PMC10311417
Seyyed Morteza Hoseini, Morteza Yousefi, Alireza Afzali-Kordmahalleh, Esmaeil Pagheh, Ali Taheri Mirghaed
Animals (Basel) 2023 Jun; 13(12): 1934. Published online 2023 Jun 9. doi: 10.3390/ani13121934
PMCID: PMC10295004
Proline-related gene expression contribute to physiological changes of East Nusa Tenggara (Indonesia) local rice cultivars during drought stress
YUSTINA CAROLINA FEBRIANTI SALSINHA, SITI NURBAITI, ALFINO SEBASTIAN,DIDIK INDRADEWA, YEKTI ASIH PURWESTRI, DIAH RACHMAWATI, BIODIVERSITAS 2022 July Volume 23, Number 7, Pages: 3573-3583
Evolutionary dynamics of SARS-CoV-2 circulating in Yogyakarta and Central Java, Indonesia: sequence analysis covering furin cleavage site (FCS) region of the spike protein
Nastiti Wijayanti, Faris Muhammad Gazali, Endah Supriyati, Mohamad Saifudin Hakim, Eggi Arguni, Marselinus Edwin Widyanto Daniwijaya, Titik Nuryastuti, Matin Nuhamunada, Rahma Nabilla, Sofia Mubarika Haryana, Tri Wibawa, Int Microbiol. 2022 Feb 14;1-10. doi: 10.1007/s10123-022-00239-8.
PMCID: PMC8853438
Detection of SARS-CoV-2 spike protein D614G mutation by qPCR-HRM analysis
Faris Muhammad Gazali, Matin Nuhamunada, Rahma Nabilla, Endah Supriyati, Mohamad Saifudin Hakim, Eggi Arguni, Edwin Widyanto Daniwijaya, Titik Nuryastuti, Sofia Mubarika Haryana, Tri Wibawa, Nastiti Wijayanti, Heliyon. 2021 Sep;7(9):e07936. doi: 10.1016/j.heliyon.2021.e07936. Epub 2021 Sep 6.
PMCID: PMC8420086
BMPR2 Editing in Fibroblast NIH3T3 using CRISPR/Cas9 Affecting BMPR2 mRNA Expression and Proliferation
Dwi Aris Agung Nugrahaningsih, Eko Purnomo, Widya Wasityastuti, Ronny Martien, Nur Arfian, Tety Hartatik, The Indonesian Biomedical Journal, 2022, Vol 14, No 1. DOI: https://doi.org/10.18585/inabj.v14i1.1724
ABScript III RT Master Mix for qPCR with gDNA Remover (RK20433)
|
ABScript III RT Master Mix for qPCR with gDNA Remover is developed based on ABScript III Reverse Transcriptase and suitable for two-step RT-qPCR detection. The 5X ABScript III RT Mix in this product contains all the reagents required for the reverse transcription reaction. The reaction protocol is simple and can be carried out quickly by adding the RNA template and H2O. The gDNA Remover Mix in this product can completely remove the genomic DNA remaining in the RNA template and make the qPCR results more accurate. The dsDNase is heat-sensitive and can be quickly and irreversibly inactivated under high temperature conditions. Therefore, it only needs one sample to be used to remove genomic DNA contamination and reverse transcription reactions in the same tube.
This product is specially optimized for qPCR. The proportionally optimized Random Primers/Oligo (dT)20VN Primer Mix enables cDNA synthesis to progress from each region of RNA transcription efficiently, which ensures the authenticity and repeatability of qPCR results to the greatest extent. Reverse transcription products are compatible with SYBR Green and probe qPCR and can be used in combination with corresponding reagents according to experimental purposes for high-performance geneexpression analysis. |
Product Feature
- Convenient operation: gDNA removal and reverse transcription can be completed in one preparation, and the entire experimental process can be completed within 20 minutes.
- gDNA removal: It can effectively remove gDNA from the sample without affecting reverse transcription efficiency;
- Universality: It has a more stable reverse transcription efficiency for genes with different abundance and GC content;
- Inhibitor tolerance: Common inhibitors introduced in RNA extraction, such as salt ions, ethanol, phenols, and SDS, have strong tolerance and better tolerance to low-quality samples.
Product Component
Product Data
1. Excellent reverse transcription efficiency and universality
|
Use RK20433 and an imported brand to reverse transcription 1 μg human K562 cell RNA and qPCR detection of 10 genes with different expression abundances.The results showed that there was high consistency between RK20433 and imported brands for high and middle abundance genes, but for low abundance genes, the Ct value of RK20433 was lower, and one gene, RK20433, was able to detect signals, while imported brands were unable to detect signals
|
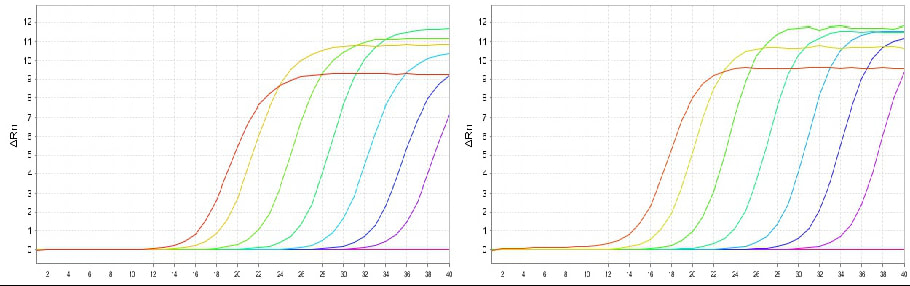
2. Wide Sample starting quantity compatibility
- Rat liver samples - RNA input amounts of 1000 ng, 100 ng, 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, H2O;
Mouse liver sample - RNA input amounts of 1000 ng, 100 ng, 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, H2O
Rat and Mouse liver tissue samples were subjected to 7 10-fold gradient dilutions of RNA, and reverse transcription and qPCR detection were performed using the gradient diluted RNA as a template. The results showed that in RK20433 can maintain extremely high reverse transcription efficiency within the concentration range of 1 μg-1 pg
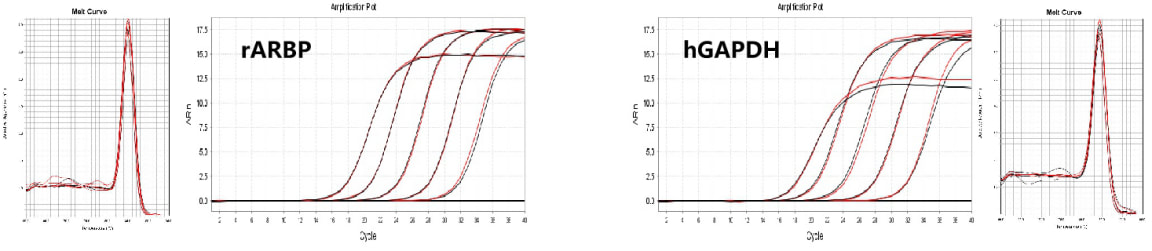
3. The effect of gDNA Remover on reverse transcription efficiency
- To detect the effect of gDNA Remover on reverse transcription efficiency, reverse transcription of human blood samples and rat tissue samples RNA was performed with RM21485 (reverse transcription component in RK20433, corresponding to the black amplification curve in the figure) and RM21486+RM21483 (reverse transcription component in RK20433+gDNA Remover component, corresponding to the red amplification curve in the figure), In a 20 μL system, 2 μL of 100 ng/μL was added for reverse transcription, and the product cDNA was diluted in a 10 fold gradient: 1X, 0.1X, 0.01X, 0001X, 0.0001X, H2O. The results showed that adding or not adding gDNA Remover components had no effect on downstream experimental results.